There persists a misconception that if things are measured in the same channel, they are the same color or “close enough”. The example that I’ll use to illustrate the potential problems with this was a panel that used BV510 and V500 as the labels for the dump channel markers, and Live/Dead Aqua for viability, which are all measured off of 405 excitation and a bandpass around 525/50. You can see in the image below that they are all slightly different:
http://www.biolegend.com/spectraanalyzer A quick aside:
On our system, we have the following detectors on our violet laser. Shown are the uncompensated signals from the 3 different dyes on single stained beads.
If you compensate based on V500, you can see the other parameters are not properly compensated in all channels:
Compensating based on BV510:
And Live/Dead Aqua:
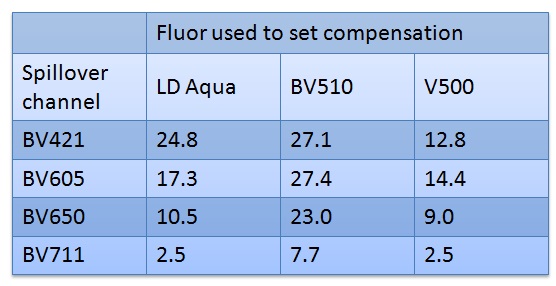
The cytometer has no way to tell WHICH dye is generating the signal, so it will not be able to correct the signals properly, but will just apply whatever values your matrix contains. The actual matrix values for the different dyes:
If you still decide to mix fluors, in this case (viability combined with a dump channel) I would use the Live/Dead dye to determine compensation. ALL cells will be stained with the L/D dye (dead cells will be brightest) so errors in compensation will affect your populations of interest. For a dump channel you are keeping the negative cells so even if compensation isn’t quite right, it should not affect your cells of interest.
Note that a similar situation can occur if you use a tandem dye for your dump channel–the tandem for each different antibody may be slightly different, so your compensation may not be correct for all markers.







I am also thankful that Biolegend has phycoerythrin listed as PE instead of R-PE, which I would never think to look for and causes me to look stupid in front of clients when I am attempting to show them the spectra… Bookmarking the Biolegend site NOW!
The visuals in your explanation are very convincing – hope folks will heed the warning!
There are a bunch of other spectraviewers linked above under Helpful Links>Spectraviewers, as not every spectraviewer (not even Biolegend’s!) has every spectra:
If there are any suggestions for others to add please send them to me!
Pingback: FACS Pain | New England Cytometry